Still images
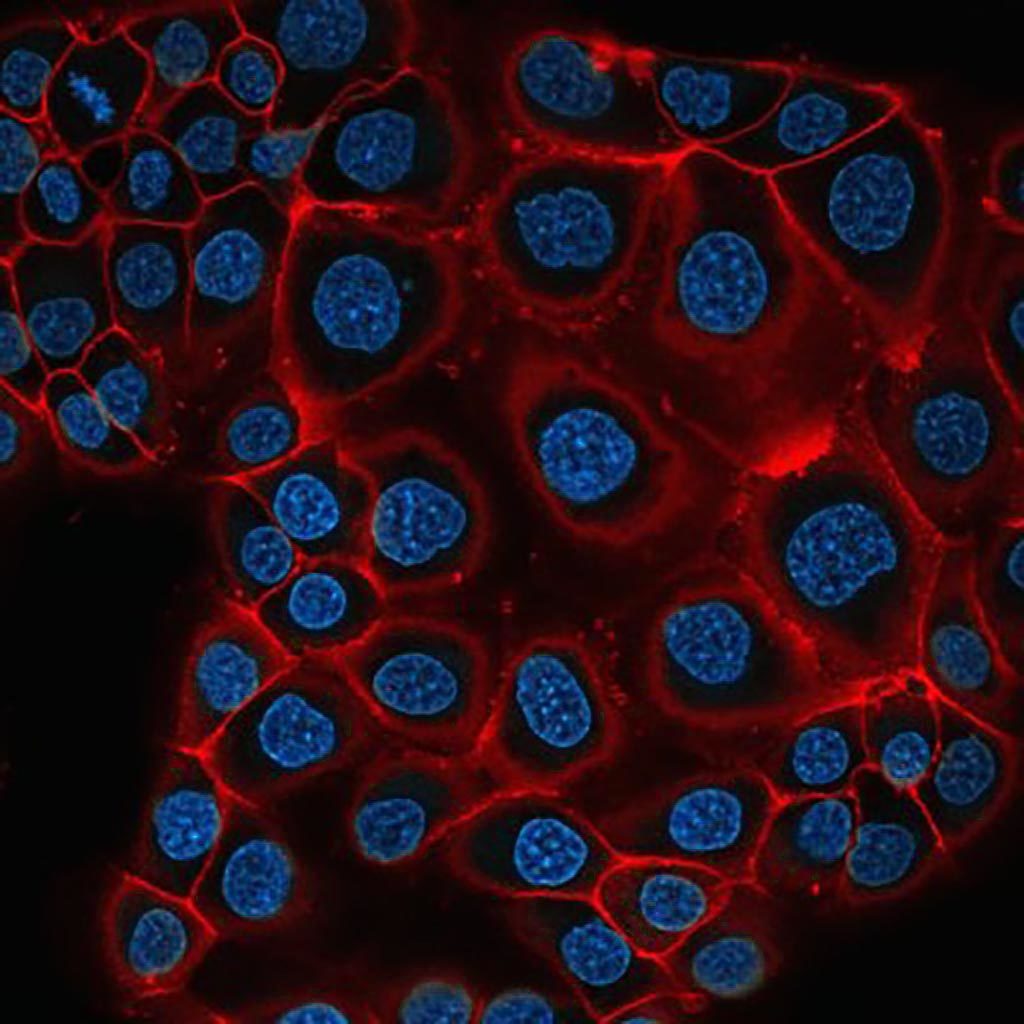
PNT2C2 an eptithelial prostate cell line stained with pKH26 (red) as a membrane marker and Hoechst (cyan) staining the nuclei. Single slice of a confocal 3D stack.
Thomas Hartjes and Gert van Cappellen
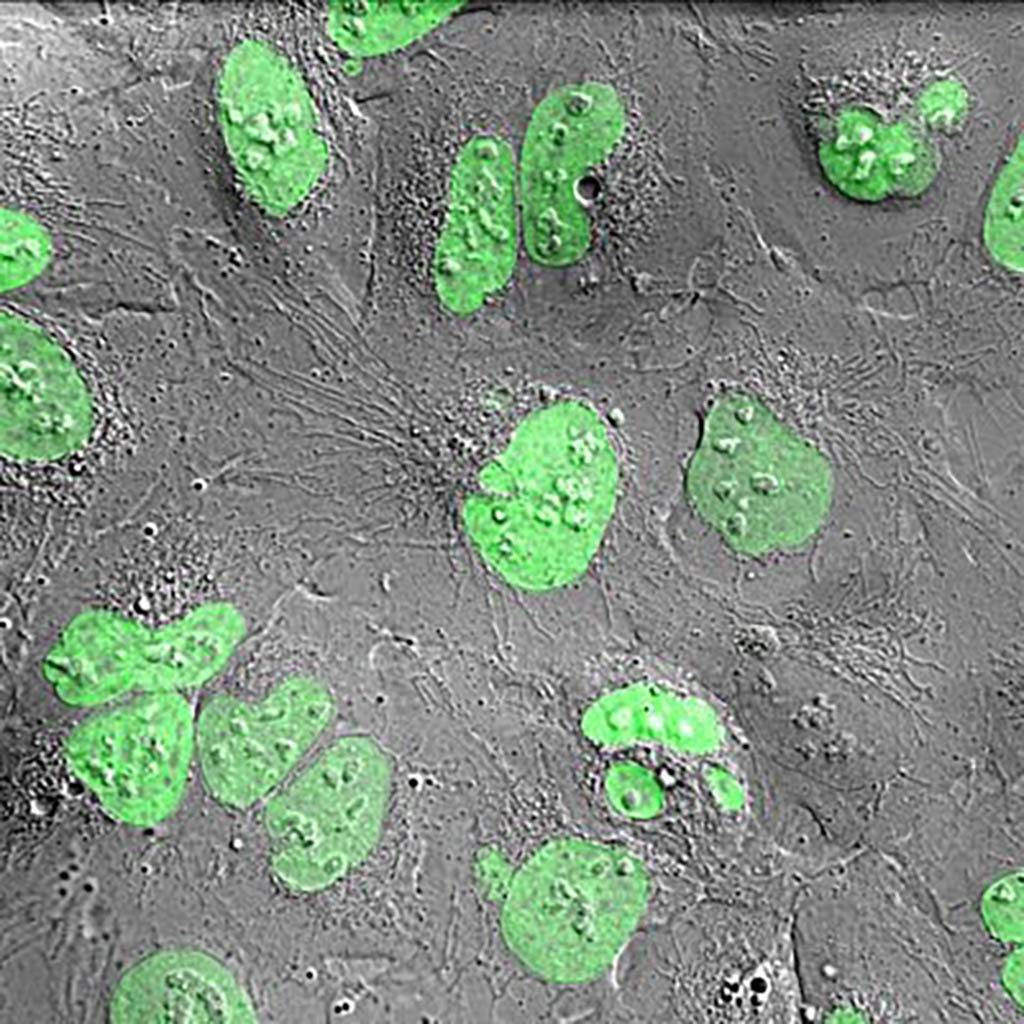
Hela cells stably transfected with Histon-2B-GFP. The confocal signal is overlaid with a Differential Interference Contrast (DIC) image which is not confocal.
Tsion Abraham and Gert van Cappellen
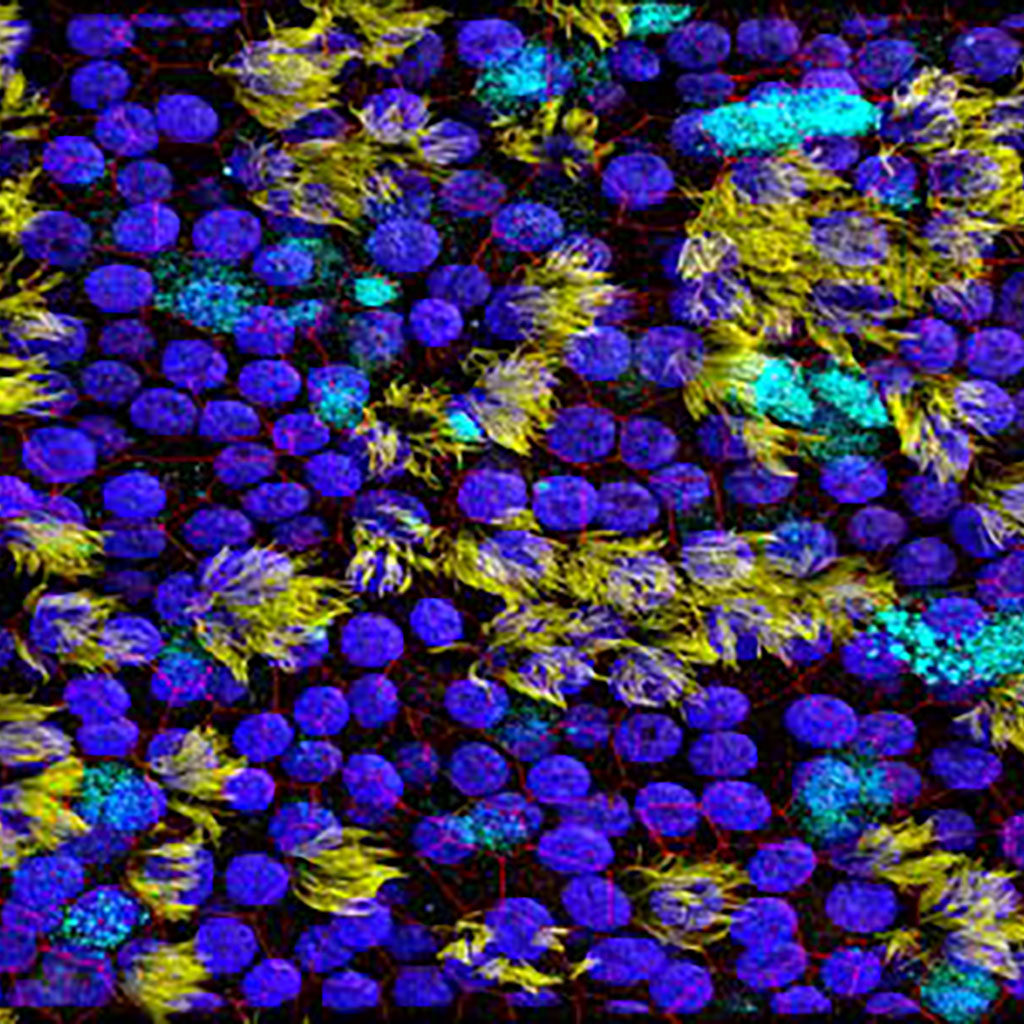
Ciliated respiratory epithelial cells (yellow) and mucus producing goblet cells (cyan), containing tight junctions (red) and nuclei (blue).
Alwin de Jong, Gert-Jan Kremers en Rik de Swart
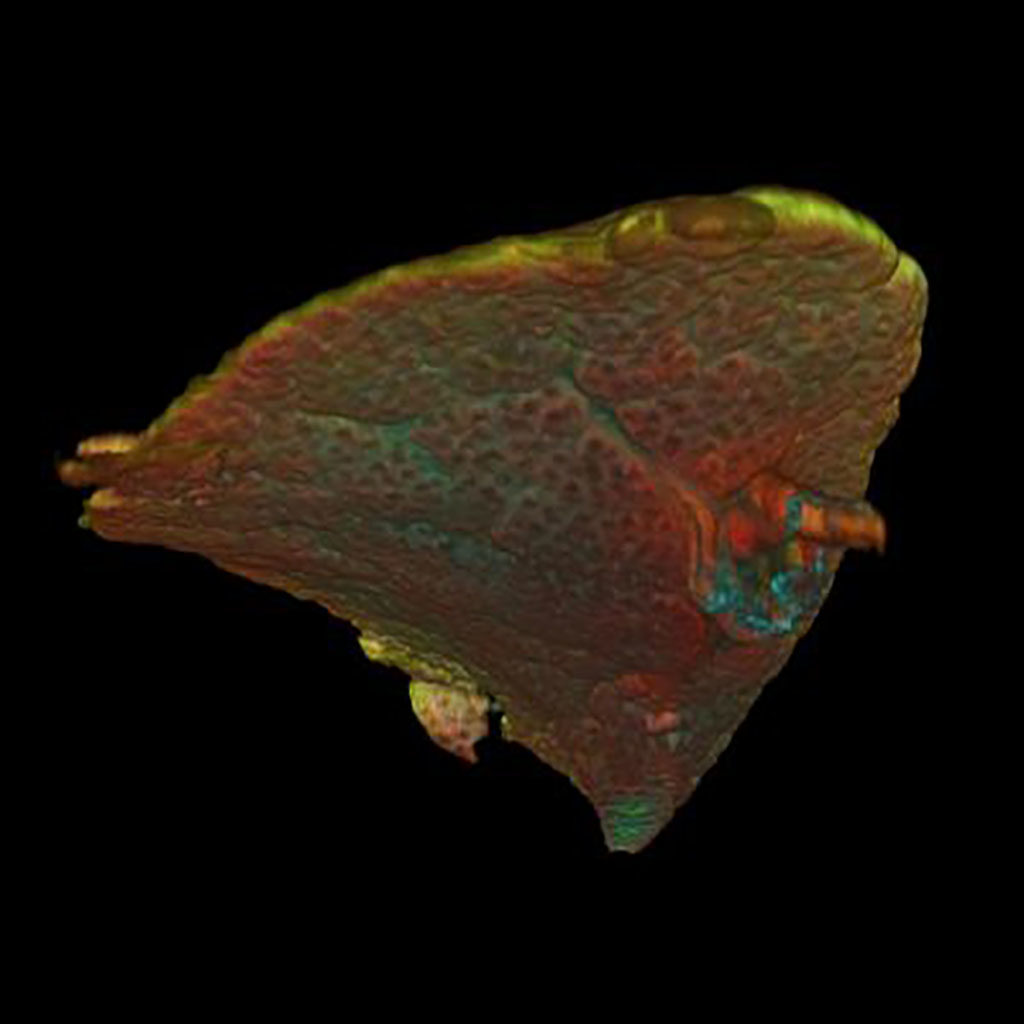
Embryonic mouse lung day E15, with stained pulmonary vessels (Green) Blood vessels (Cyan) and special cells (Red). Confocal Microscope, volume rendering.
Image Petra Burgisser (Heleen Kool, Robbert Rottier)
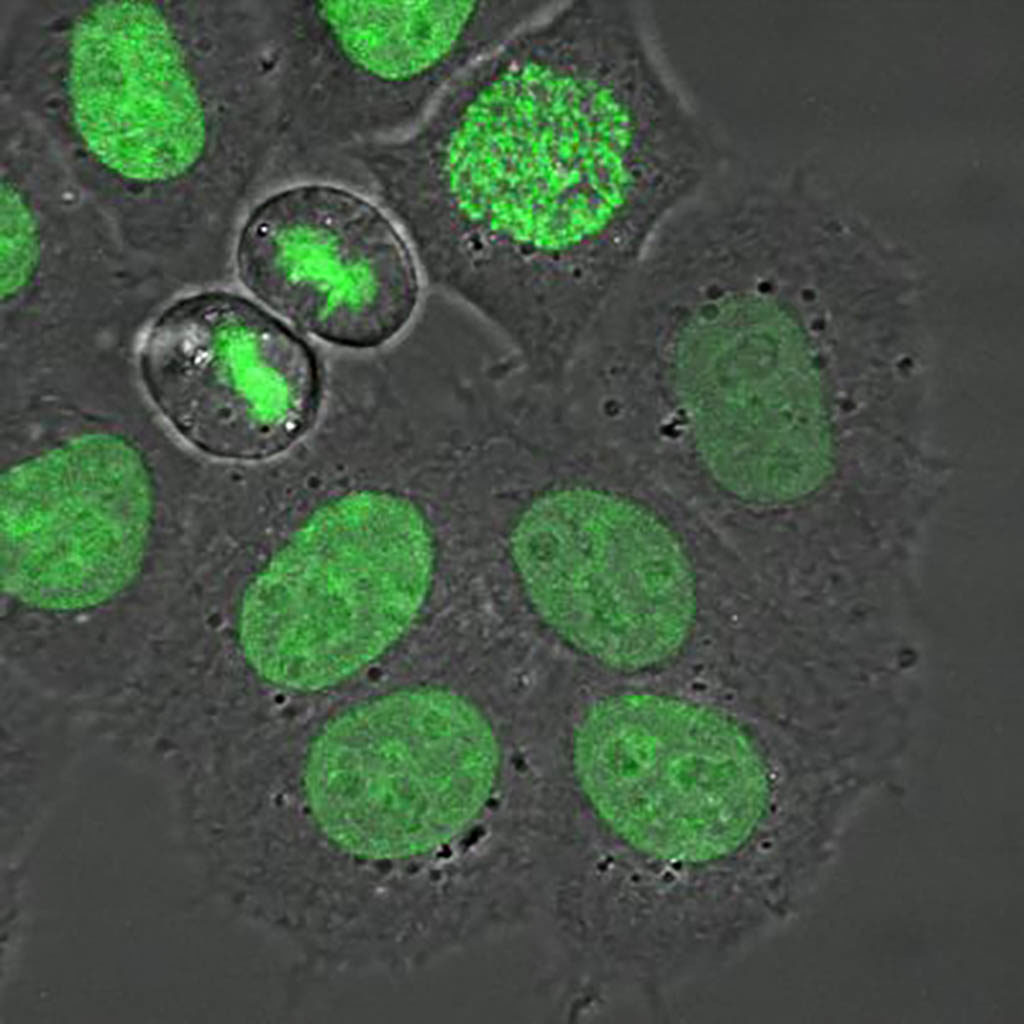
Dividing H2B-GFP hela cells imaged with a confocal microscope using a transmission and fluorescence channel. Exitation 488 nm and emission BP 500-550 nm.
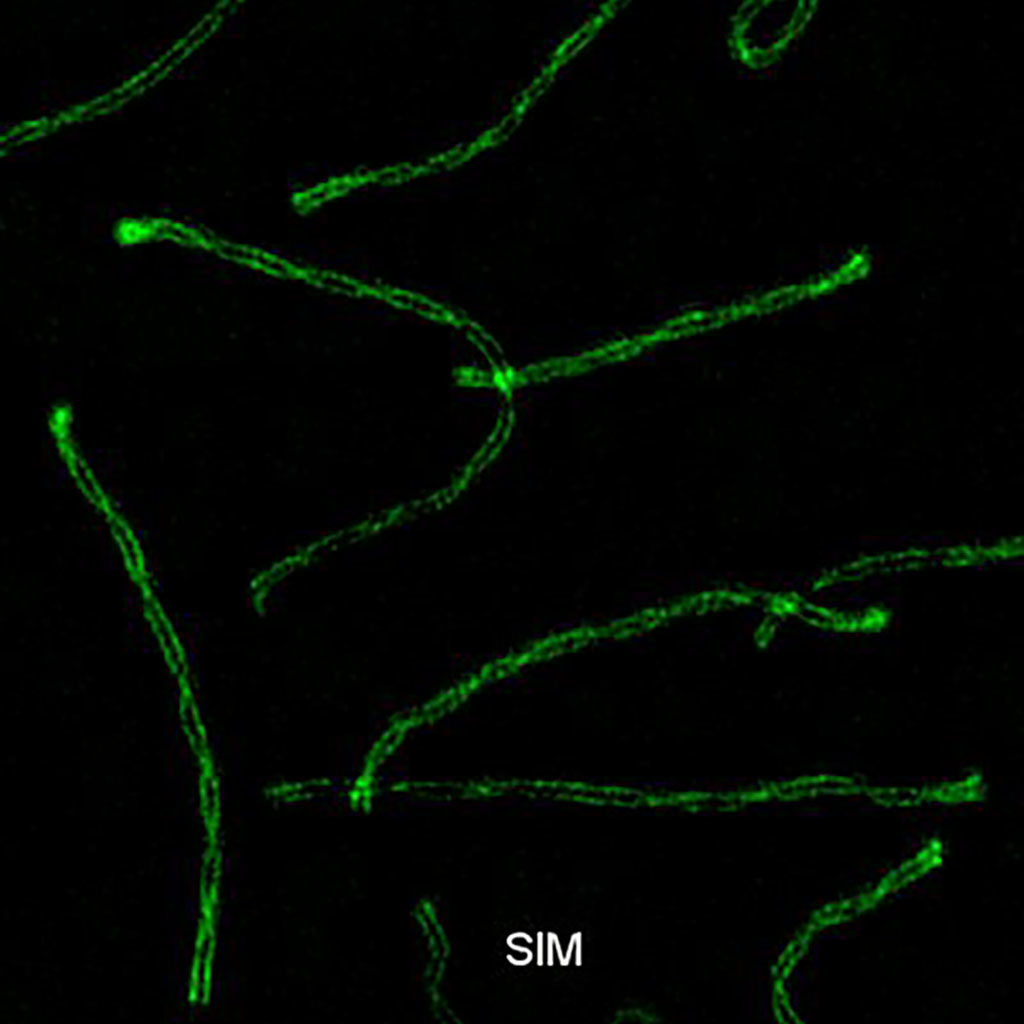
Synaptonemal complex during meiosis in spermatocytes.
Structured illumination microscopy (SIM) SYCP3-Alexa 488. Exitation 488 nm. Emission BP 500 – 585 nm.
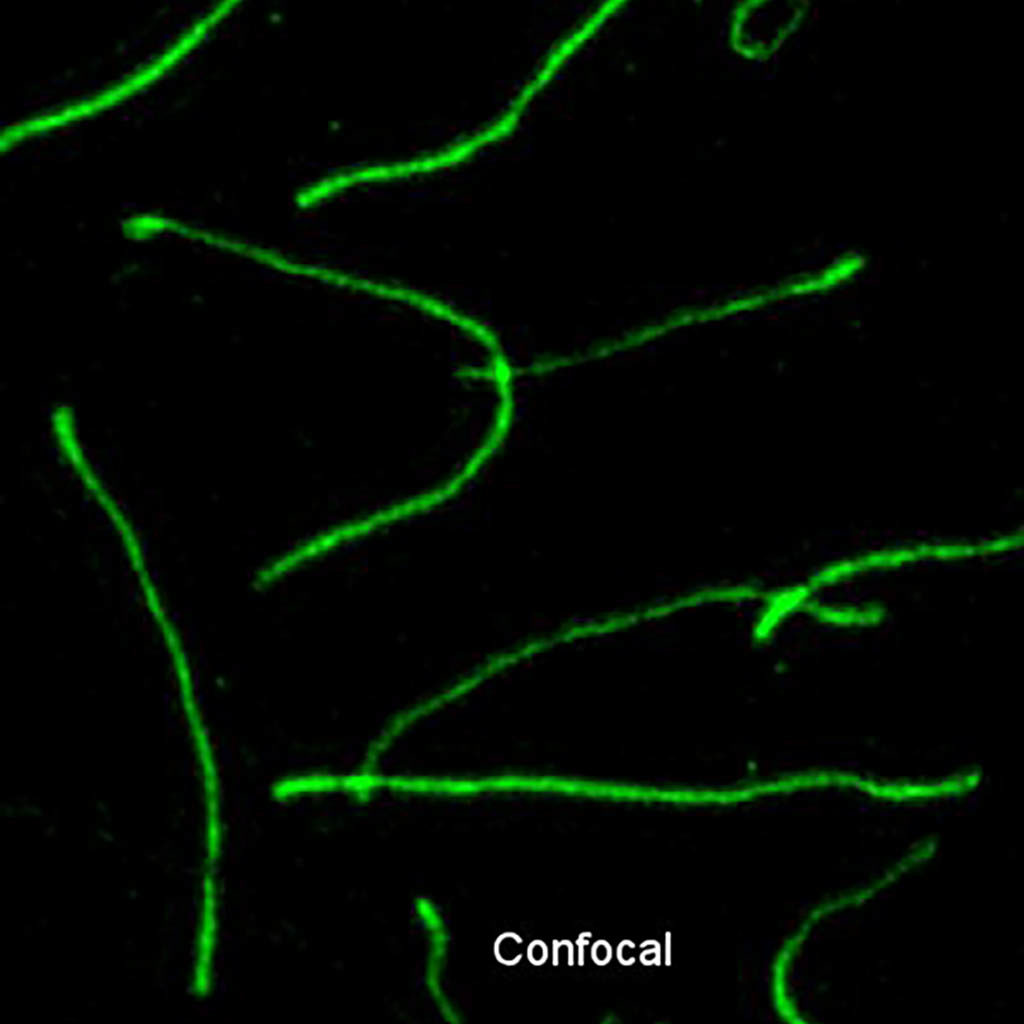
Synaptonemal complex during meiosis in spermatocytes.
Structured illumination microscopy (SIM) and Confocal SYCP3-Alexa 488. Exitation 488 nm. Emission BP 500 – 585 nm zoom in.
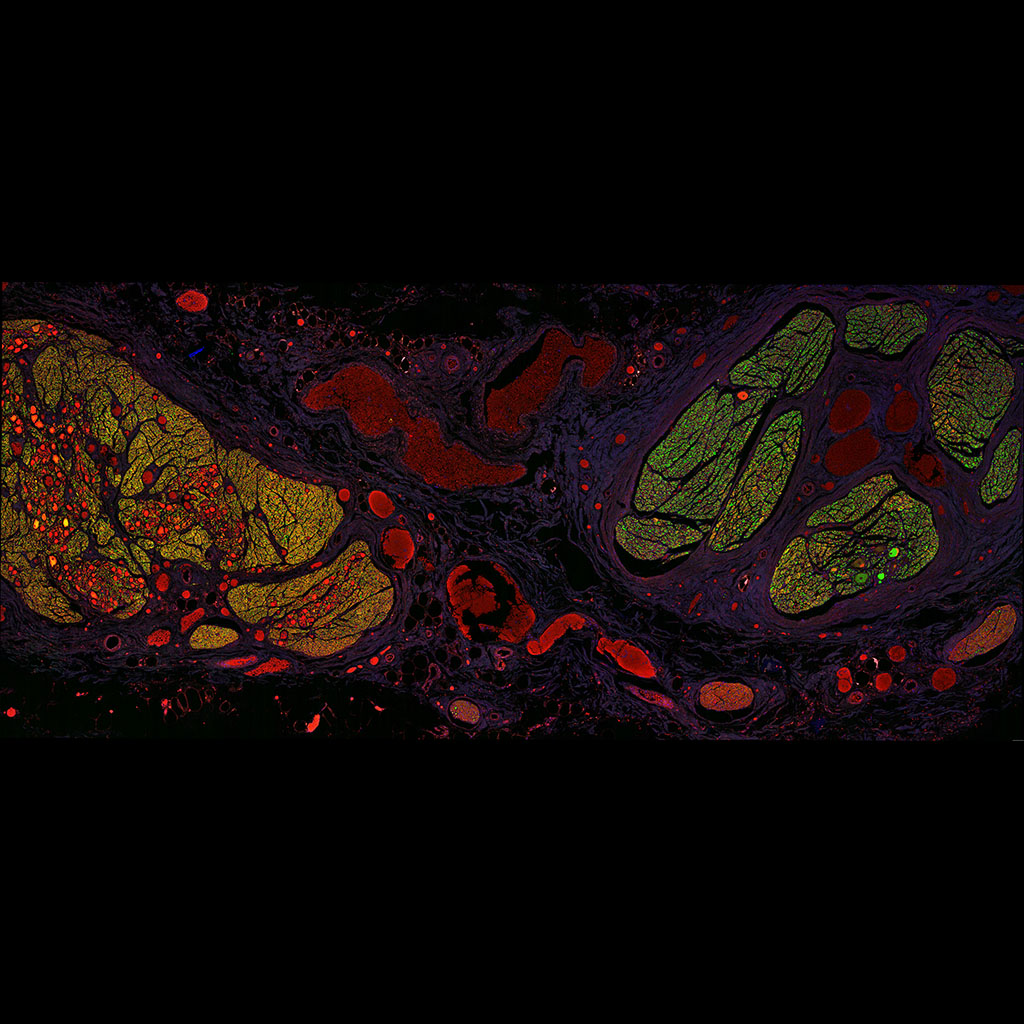
Vagus nerve (right) and truncus sympathaticus (left) approximately at the the superior cervical ganglion, from an embalmed body that was made available to Erasmus MC. The structures between the two nerves are blood vessels (so it is a transverse section). Tile scan Stellaris
Lara Orlandini (Tom Ruigrok)
3D movies
3D stack of anaphase of prostate epithelial cells (PNT2C2) stained with Hoechst (dna, blue) and PKH (membrane of the cell, green). The sample was made by Thomas Hartjes and imaged on a confocal microscope by Nanobiology bachelor students, during the Nano-microscopy practicals.
The students were supervised by Gert van Cappellen
Hela cell with Histon2B-GFP going through mitosis. Zeiss LSM510 confocal microscope projected with the Huygens (SVI) SFP volume renderer.
Image and movie made by Gert van Cappellen
Reconstructed structured illumination microscopy (SIM) image of a fluorescent nuclear track detector (Al2O3:C,Mg) after exposure with an Am-241 alpha particle source.
Jasper Kouwenberg and Johan Slotman J Microsc. 2018 Feb 2. (PMID: 29393521)
4Pi microscopy of prostate cancer-derived extracellular vesicles uptake by prostate epithelial cells imaged. Cells are labeled with PKH67 (membrane, green) and SirDNA (nucleus, blue) and EVs are labeled with PKH26 (red).
Thomas Hartjes, Gert van Cappellen, Martin van Royen
3 interphase and one prophase Hep3B cells stained with Hoechst (DNA). Confocal microscope (Zeiss LSM700) excitation 405 nm, Hoechst 34580, pinhole 0.5 AU. Deconvolved with Huygens (SVI), volume rendering ImageJ 3D viewer plugin (Benjamin Schmid).
Image made by Alex Nigg
3 interphase and one prophase Hep3B cells stained with Propidium iodide (DNA and RNA). Confocal microscope (Zeiss LSM700) excitation 555 nm, pinhole 0.5 AU. The PI DNA signal was removed using the Hoechst signal. Deconvolved with Huygens (SVI), volume rendering ImageJ 3D viewer plugin (Benjamin Schmid).
Image made by Alex Nigg
3 interphase and one prophase Hep3B cells stained with Propidium iodide (RNA) in Red and Hoechst (DNA) in gray. Confocal microscope (Zeiss LSM700) excitation 405 and 555 nm, in separate tracks, pinhole 0.5 AU. The PI DNA signal was removed using the Hoechst signal. Deconvolved with Huygens (SVI), volume rendering ImageJ 3D viewer plugin (Benjamin Schmid).
Image made by Alex Nigg
Prophase nucleus of Hep3B cell stained with Hoechst. Confocal microscope (Zeiss LSM700) excitation 405 nm, Hoechst 34580, pinhole 0.5 AU. Deconvolved with Huygens (SVI), volume rendering ImageJ 3D viewer plugin (Benjamin Schmid).
Image made by Alex Nigg
Interphase and prophase of Hep3B cell nuclei cell stained with Hoechst. Confocal microscope (Zeiss LSM700) excitation 405 nm, Hoechst 34580, pinhole 0.5 AU. Deconvolved with Huygens (SVI), volume rendering ImageJ 3D viewer plugin (Benjamin Schmid).
Image made by Alex Nigg
3D connections between PNT2C2 (normal human prostate epithelium) labeled with PKH67 (membrane, green) and Sytox (nucleus, blue) imaged with a 4Pi microscope
Sharzad Pouyanrad, Alex Nigg and Martin van Royen
Timelapse
Dividing U2OS H2B-mMaple cells imaged with a confocal microscope every 5 seconds. The top right cell goes from prophase trough metaphase to anaphase. Also two other cells divide.
Adriaan Houtsmuller
Dividing H2B-GFP hela cells imaged with a confocal microscope using a transmission and fluorescence channel. Exitation 488 nm and emission BP 500-550 nm.
Plus end binding protein Clip170-GFP in 3T3 cells. Confocal microscope excitation 488 nm, emission BP500-550 nm. Time lapse of 49 time points, every 2 seconds, total 98 seconds.
Tatiana Stepanova, group Niels Galjart, Gert van Cappellen
Birth of a Haemayopoietic Stem Cell in the Aorta of an embryonic mouse, full field of view. Confocal microscope, 40x dry lens (n.a. 0.6) pinhole 2 airy units, Z-range 125 µm in 5 µm steps, pixelsize 292×292 µm. Ext 488 (Ly6a-GFP) Em BP 500-550; Ext 561 (CD31-PE) Em 570-620. XYZ drift correction with the Stackreg plugin (P. Thévenaz, U.E. Ruttimann, M. Unser) in ImageJ (Wayne Rasband). Deconvolved with Huygens (SVI) deconvolution software.
Jean-Charles Boisset, Catherine Robin, Gert van Cappellen
Birth of a Haemayopoietic Stem Cell in the Aorta of an embryonic mouse, cropped and rotated field of view. Confocal microscope, 40x dry lens (n.a. 0.6) pinhole 2 airy units, Z-range 125 µm in 5 µm steps, pixelsize 292×292 µm. Ext 488 (Ly6a-GFP) Em BP 500-550; Ext 561 (CD31-PE) Em 570-620. XYZ drift correction with the Stackreg plugin (P. Thévenaz, U.E. Ruttimann, M. Unser) in ImageJ (Wayne Rasband). Movie was showed in the 20h NOS journal. Deconvolved with Huygens (SVI) deconvolution software.
Jean-Charles Boisset, Catherine Robin, Gert van Cappellen
Prostate
3D Normal prostate_250 micrometer:
Three-dimensional imaging of normal epithelial prostate gland. Deconvoluted nuclear RedDot staining (approx. 250 micrometer).
3D Prostate cancer Gleason 3 glandular_250 micrometer:
Three-dimensional imaging of glandular prostate adenocarcinoma, Gleason score 3+3=6. The tumor is forming simple tubules with sporadic branching and interconnections. Deconvoluted nuclear RedDot staining (approx. 250 micrometer).
3D Prostate cancer Gleason 4 fused_250 micrometer:
Three-dimensional imaging of prostate adenocarcinoma, Gleason score 3+4=7. The tumor forms simple tubules (Gleason grade 3) and fused sheets (Gleason grade 4). Deconvoluted nuclear RedDot staining (approx. 250 micrometer).
Z-stack Prostate cancer Gleason 4 fused E-cad_900 micrometer:
Z-stack image of prostate adenocarcinoma, Gleason score 4+3=7. Extensive fusion of glands and sheets (Gleason grade 4). Nuclear RedDot staining and membranous E-Cadherin directly labelled with Alexa 488 (approx. 900 micrometer).
Plusend
EB3 Plusends in ES Cells:
ES cells expressing EB3-GFP were imaged with the Luxendo InVi fluorescence light sheet microscope at 50 frames per second. Note the movement of EB3-GFP “comets” (representing the ends of growing microtubules). ES cells have many dynamic microtubules, most of which originate from the centrosome in the middle of the cell.

